Example report of OUKS
This R Markdown document was provided as an example to reproduce the OUKS code script. Fig. 4 from article can be reproduced with the following code. Preliminarily download “xcms after IPO MVI QC-XGB filter repeats annot+filtr LMM adj KEGG.csv” and “8 peaks.csv” files into your working directory (for example: “D:/OUKS/”).
For quick access to files:
ds <- as.data.frame(fread("https://raw.githubusercontent.com/plyush1993/OUKS/main/Datasets%20(csv)/8%20peaks.csv"))
ds2 <- as.data.frame(fread("https://raw.githubusercontent.com/plyush1993/OUKS/main/Datasets%20(csv)/xcms%20after%20IPO%20MVI%20QC-XGB%20filter%20repeats%20annot%2Bfiltr%20LMM%20adj%20KEGG.csv"))
Prepare environment
First, set the folder for the working directory and load the packages.
setwd("D:/OUKS/")
library(data.table)
library(factoextra)
library(FactoMineR)
library(dendextend)
library(rafalib)
library(pROC)
library(parallel)
library(doParallel)
library(grid)
library(caret)
library(dplyr)
library(MKinfer)
library(limma)
library(ggplot2)
library(ggsci)
Load datasets
ds <- as.data.frame(fread(input = "8 peaks.csv", header=T))
rownames(ds) <- ds[,1]
ds <- ds[,-1]
colnames(ds)[1] <-"Label"
ds[,-1] <- sapply(ds[,-1], as.numeric)
ds$Label <- as.factor(ds$Label)
ds2 <- as.data.frame(fread(input = "xcms after IPO MVI QC-XGB filter repeats annot+filtr LMM adj KEGG.csv", header=T))
ds2 <- ds2[-c(1:12),]
rownames(ds2) <- ds2[,5]
ds2 <- ds2[,-c(1,3:5)]
ds2[,-1] <- sapply(ds2[,-1], as.numeric)
ds2$Label <- as.factor(ds2$Label)
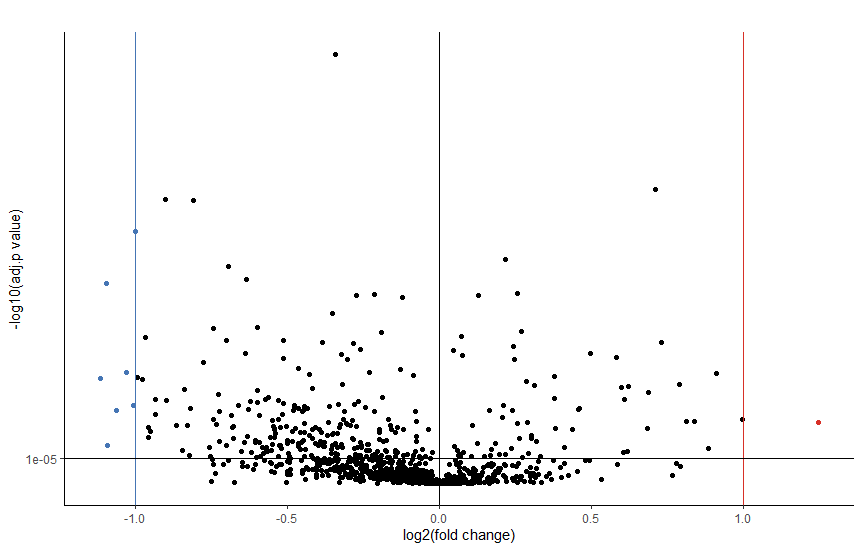
Plot volcano plot
ds_log <- as.data.frame(log2(ds2[,-1]))
ds_log <- cbind(Label = ds2[,1], ds_log)
FOLD.CHANGE <- function(data) {
ds_log_subsets <- lapply(1:length(unique(data[,1])), function(y) filter(data[,-1], data$Label == unique(data[,1])[y]))
mean_r_l <- lapply(1:length(ds_log_subsets), function(y) apply(ds_log_subsets[[y]], 2, mean, na.rm = T))
foldchange <- mean_r_l[[1]] - mean_r_l[[2]]
fc_res <- as.data.frame(foldchange)
return(fc_res)
}
fc_res <- FOLD.CHANGE(ds_log)
foldchange <- as.numeric(fc_res$foldchange)
mdl_mtrx <- model.matrix(~Label, ds2)
lmf <- lmFit(t(ds2[,-1]), method = "robust", design = mdl_mtrx, maxit = 1000)
efit <- eBayes(lmf)
tableTop <- topTable(efit, coef = 2, adjust = "BH", number = ncol(ds2), sort.by = "none")
pval <- as.numeric(tableTop$adj.P.Val)
f <- volcano(foldchange, pval, effect.low = -1.0, effect.high = 1.0, sig.level = 0.00001,
xlab = "log2(fold change)", ylab = "-log10(adj.p value)", title = "") + theme_classic() + theme(legend.position="none")
f
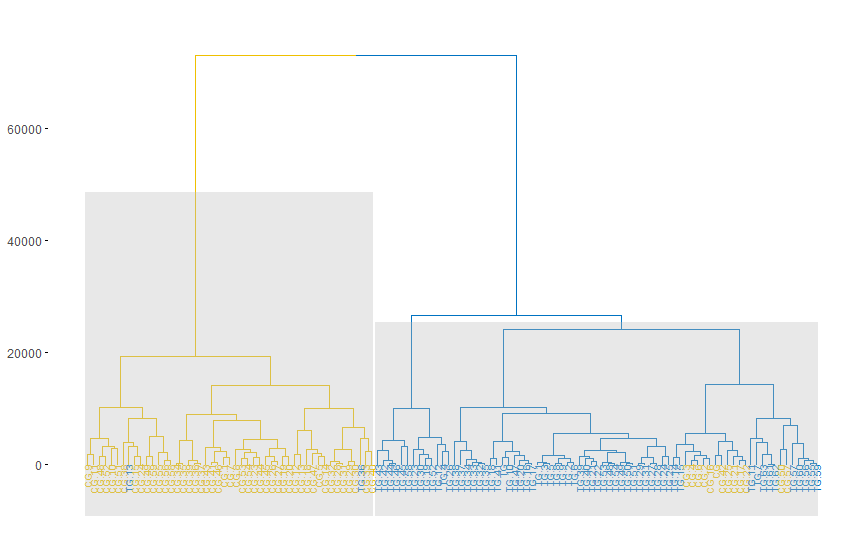
HCA plot
base1 <- ds
mtrx1 <- ds[,-1]
grp1 <- as.character(base1[,1])
k <- length(unique(grp1))
Cols = function(vec, ord){
cols = pal_jco(palette = c("default"), alpha = 1)(length(unique(vec)))
return(cols[as.fumeric(vec)[ord]])}
mtrx1_1 <- mtrx1
rownames(mtrx1_1) = make.names(grp1, unique=TRUE)
res.dist1 <- dist(mtrx1_1, method = "manhattan")
res.hc1 <- hclust(d = res.dist1, method = "ward.D2")
b <- fviz_dend(res.hc1, k = k,
cex = 0.55,
k_colors = unique(Cols(grp1,res.hc1$order)),
color_labels_by_k = F,
label_cols = Cols(grp1,res.hc1$order),
rect = T,
rect_fill = T,
horiz = F,
lwd = 0.7,
show_labels = T,
main = "",
ylab = "")
b
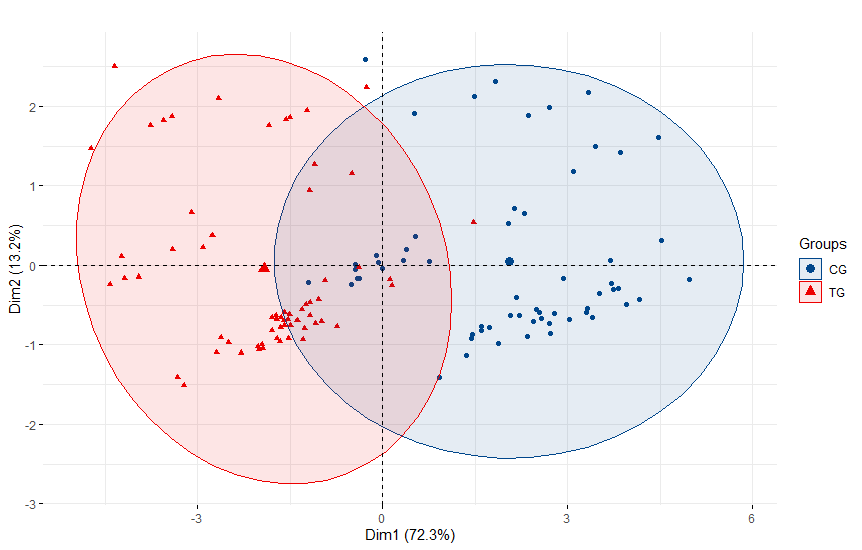
PCA plot
base1 <- ds
mtrx1 <- ds[,-1]
grp1 <- as.character(base1[,1])
palette_pca <- "lancet"
pca.ds1 <- PCA(mtrx1, scale.unit = T, graph = F)
a <- fviz_pca_ind(pca.ds1,
title = "",
geom.ind = "point",
col.ind = grp1,
palette = palette_pca,
addEllipses = T,
legend.title = "Groups")
a
ROC curve
# start parallel processing
fc <- as.numeric(detectCores(logical = T))
cl <- makePSOCKcluster(fc-1)
registerDoParallel(cl)
# cross-validation
set.seed(1234)
trainIndex <- createDataPartition(ds$Label, p = 0.8, list = F, times = 1)
dsTrain <- ds[ trainIndex,]
dsValid <- ds[-trainIndex,]
trainControl <- trainControl(method="repeatedcv", number=10, repeats=10, classProbs = T)
metric <- "Accuracy"
# machine learning
set.seed(1234)
fit.cl <- train(Label~., data=dsTrain, method="svmRadial", metric=metric, trControl=trainControl, tuneLength = 10)
predicted.classes <- predict(fit.cl, newdata=dsValid)
probabilities <- predict(fit.cl, newdata=dsValid, type = "prob")[,1]
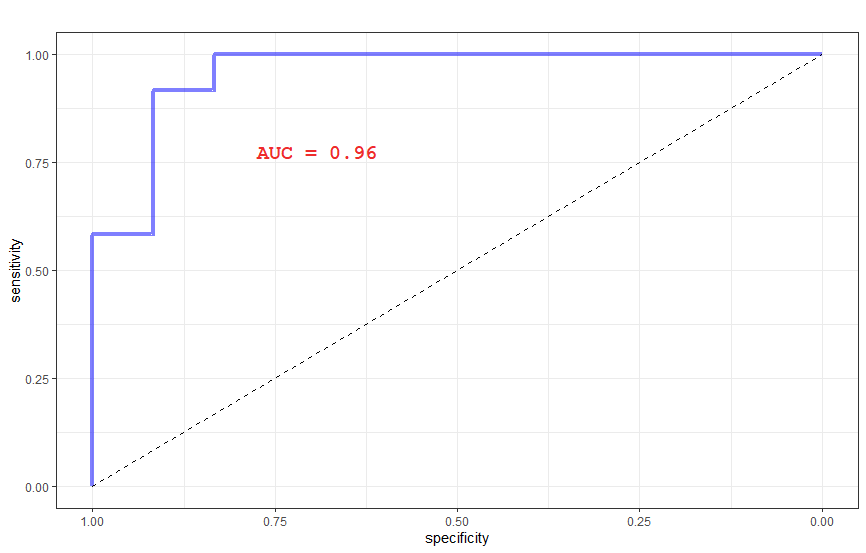
# ROC curve
res.roc <- roc(dsValid$Label, probabilities, levels = levels(dsValid$Label))
auroc <- round(as.numeric(auc(res.roc)),2)
grob <- grobTree(textGrob(paste0("AUC = ", auroc), x=0.25, y=0.75, hjust=0,
gp=gpar(col="firebrick2", fontsize=15, fontface=11)))
c <- ggroc(res.roc, alpha = 0.5, colour = "blue1", linetype = 1, size = 1.5) +theme_bw() + ggtitle("") +
geom_segment(aes(x = 1, xend = 0, y = 0, yend = 1), color="black", linetype="dashed") + annotation_custom(grob)
c
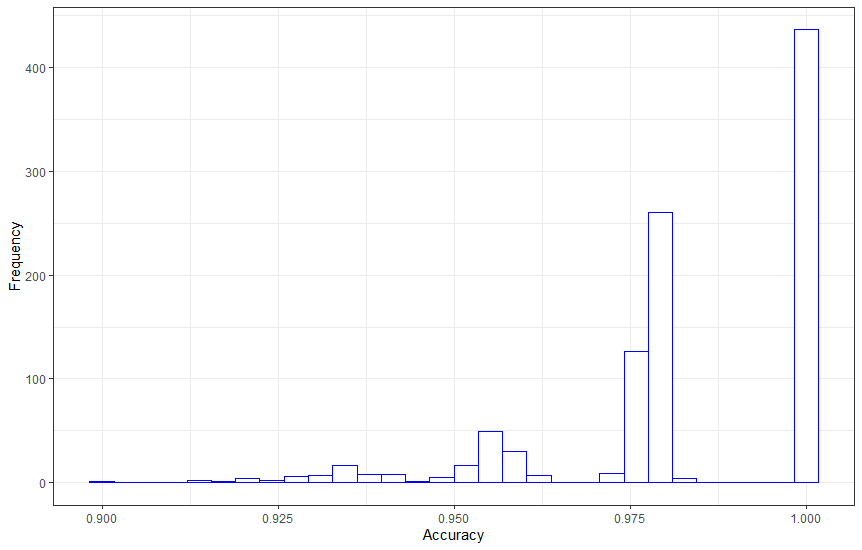
bootstrap histogram
# start parallel processing
fc <- as.numeric(detectCores(logical = T))
cl <- makePSOCKcluster(fc-1)
registerDoParallel(cl)
# bootstrap
set.seed(1234)
trainControl <-trainControl(method="boot", number=1000)
metric <- "Accuracy"
# machine learning
set.seed(1234)
fit.cl <- train(Label~., data=ds, method="svmRadial", metric=metric, trControl=trainControl)
results <- resamples(list(svm=fit.cl, svm1=fit.cl), trControl = trainControl, metric=metric)
# histogram
d <-ggplot(results$values, aes(x=results$values[,2])) +
geom_histogram(colour="blue", fill="white") + theme_bw() + xlab("Accuracy") + ylab("Frequency")
d